Dr Rajapandiyan Panneerselvam, Assistant Professor, Department of Chemistry, and his team have developed a method using a portable Raman spectrometer to quickly identify six common pathogenic Vibrio species that can contaminate seafood. His latest research paper Intelligent convolution neural network-assisted SERS to realise highly accurate identification of six pathogenic Vibrio, has been published in the Q1 Nature Index journal Chemical Communications, having an Impact Factor of 6.0.
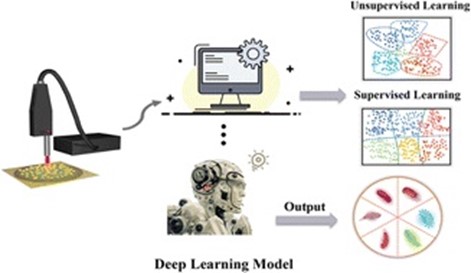 By using gold-silver nanoparticles, the study was able to accurately detect these harmful microorganisms. The new deep learning model called a convolutional neural network (CNN), outperformed traditional machine learning methods with a classification accuracy of 99.7%. The entire identification process only took 15 minutes. The researchers also discovered that the Raman signals emitted by Vibrio species are similar to signals from certain substances found in purine degradation, such as uric acid and adenine. This knowledge helps them explain why different Vibrio species produce slightly different Raman signals. Overall, the CNN-assisted Raman spectroscopy method offers a fast and accurate way to diagnose and identify harmful microorganisms responsible for food contamination.
By using gold-silver nanoparticles, the study was able to accurately detect these harmful microorganisms. The new deep learning model called a convolutional neural network (CNN), outperformed traditional machine learning methods with a classification accuracy of 99.7%. The entire identification process only took 15 minutes. The researchers also discovered that the Raman signals emitted by Vibrio species are similar to signals from certain substances found in purine degradation, such as uric acid and adenine. This knowledge helps them explain why different Vibrio species produce slightly different Raman signals. Overall, the CNN-assisted Raman spectroscopy method offers a fast and accurate way to diagnose and identify harmful microorganisms responsible for food contamination.
Abstract
The utilisation of label-free Surface-Enhanced Raman Spectroscopy (SERS) technology enabled a comprehensive analysis of the connection between Raman signals emitted by pathogenic Vibrio microorganisms and purine metabolites. Through extensive research, a sophisticated Convolutional Neural Network (CNN) model was developed, demonstrating exceptional performance with an accuracy rate of 99.7% in the rapid identification of six common pathogenic Vibrio species within a mere 15-minute timeframe. This breakthrough offers a groundbreaking approach to pathogen identification, introducing a novel and efficient method to the field.
Practical Implementation of the Research
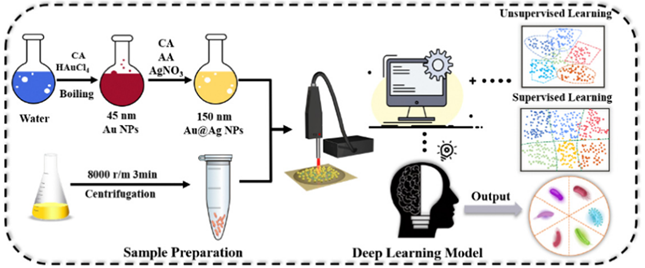 The practical implementation of label-free SERS technology combined with a deep learning CNN model enables rapid and accurate identification of pathogenic Vibrio microorganisms. This has important social implications, including improving public health and safety by quickly identifying and controlling outbreaks, enhancing food safety measures, and enabling real-time pathogen detection in resource-limited areas. The method’s speed and accuracy contribute to more informed decision-making, mitigating the spread of infectious diseases and ultimately creating a safer society.
The practical implementation of label-free SERS technology combined with a deep learning CNN model enables rapid and accurate identification of pathogenic Vibrio microorganisms. This has important social implications, including improving public health and safety by quickly identifying and controlling outbreaks, enhancing food safety measures, and enabling real-time pathogen detection in resource-limited areas. The method’s speed and accuracy contribute to more informed decision-making, mitigating the spread of infectious diseases and ultimately creating a safer society.
Future Research Plans
Moving forward, future work in the field of label-free SERS technology for pathogen identification could focus on expanding the coverage to include a wider range of Vibrio species, increasing the diversity of the dataset used for training, conducting rigorous cross-validation and external validation studies, exploring integration with portable SERS devices for on-site detection, optimising the deep learning model for speed and efficiency, and investigating clinical and environmental applications. By pursuing these avenues, the research can further enhance the versatility, reliability, and real-world applicability of the method, leading to improved methods for rapid and accurate pathogen identification in various domains.
Collaborations
- Dr Jianfeng Li (College of Materials, State Key Laboratory for Physical Chemistry of Solid Surfaces, College of Chemistry and Chemical Engineering, College of Energy, School of Aerospace Engineering, Xiamen University, Xiamen 361005, China)
- Dr Lin Zhang (State Key Laboratory of NBC Protection for Civilian, Beijing 102205, China)
- Dr Zehui Chen (Xiamen City Center for Disease Control and Prevention, Xiamen 361005, China)

