 “Research is seeing what everybody else has seen and thinking what nobody else has thought.”
“Research is seeing what everybody else has seen and thinking what nobody else has thought.”
– Albert Szent-Györgyi
“Fully Automated Quality Control of Rigid and Affine Registrations of T1w and T2w MRI in Big Data using Machine Learning” is the latest research paper published by Dr Sudhakar Tummala, Assistant Professor, Electronics and Communication Engineering, SRM University-AP in ‘Computers in Biology and Medicine Journal’ having an Impact Factor of 4.6.
Abstract of the paper:
Background: Magnetic resonance imaging (MRI)-based morphometry and relaxometry are proven methods for the structural assessment of the human brain in several neurological disorders. These procedures are generally based on T1-weighted (T1w) and/or T2-weighted (T2w) MRI scans, and rigid and affine registrations to a standard template(s) are essential steps in such studies. Therefore, a fully automatic quality control (QC) of these registrations is necessary for big data scenarios to ensure that they are suitable for subsequent processing.
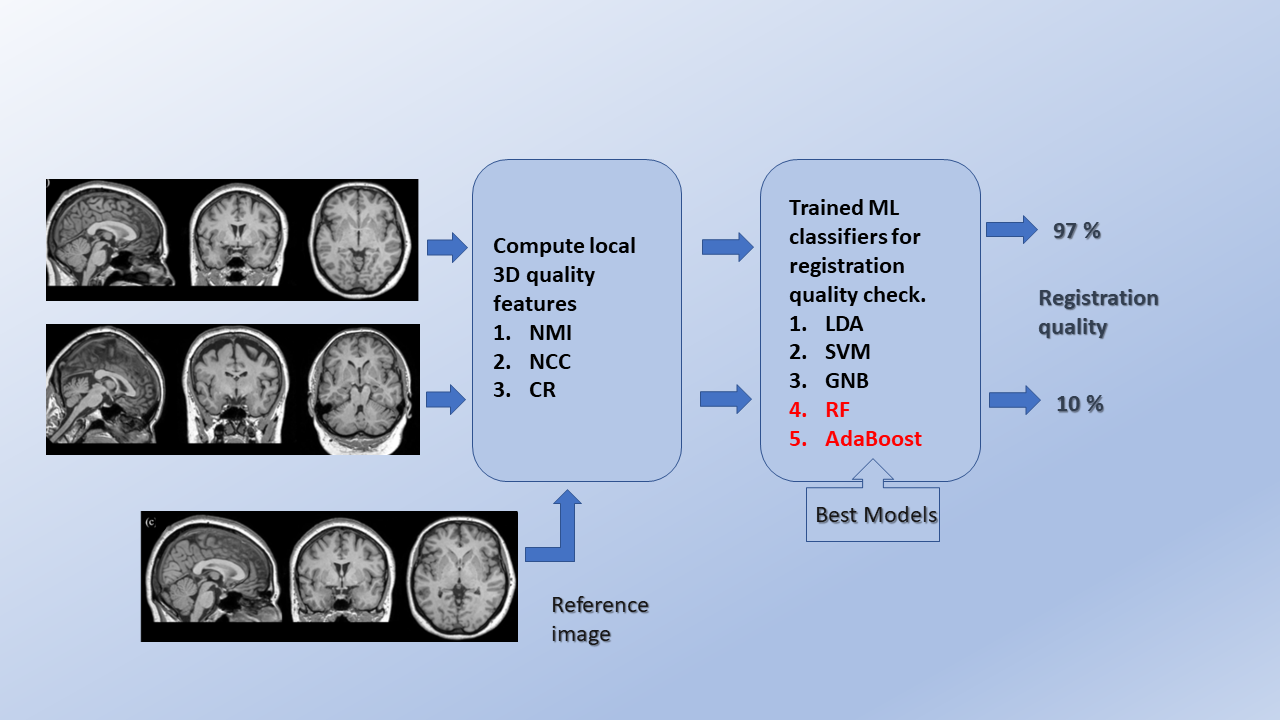
Method: A supervised machine learning (ML) framework is proposed by computing similarity metrics such as normalized cross-correlation, normalized mutual information, and correlation ratio locally. We have used these as candidate features for cross-validation and testing of different ML classifiers. For 5-fold repeated stratified grid search cross-validation, 400 correctly aligned, 2000 randomly generated misaligned images were used from the human connectome project young adult (HCP-YA) dataset. To test the cross-validated models, the datasets from autism brain imaging data exchange (ABIDE I) and information eXtraction from images (IXI) were used.
Results: The ensemble classifiers, random forest, and AdaBoost yielded the best performance with F1 scores, balanced accuracies, and Matthews correlation coefficients in the range of 0.95-1.00 during cross-validation. The predictive accuracies reached 0.99 on Test set #1 (ABIDE I), 0.99 without and 0.96 with noise on Test set #2 (IXI, stratified w.r.t scanner vendor and field strength).
Conclusions: The cross-validated and tested ML models could be used for QC of both T1w and T2w rigid and affine registrations in large-scale MRI studies.
Medical imaging is basically a method to see inside the body non-invasively. Magnetic resonance imaging (MRI) is one of the medical imaging modalities to see inside the body. MRI works on the principles of nuclear magnetic resonance. T1-weighted (T1w) and T2-weighted (T2w) MRI can enable us to see the brain without opening the skull and to monitor brain structural changes that occur in several medical and neurological conditions. It is important to monitor these structural changes over time to understand the disease progression. For example, in Alzheimer’s disease, it is important to monitor the hippocampus (memory storage centre) Recently, due to the advances in computational power such as high-performance GPUs and the availability of publicly accessible big data MRI, scientists around the world now conducting big data research studies. In group-based big data studies, for fair comparison of the brain between healthy and diseased individuals, it is necessary that the brain MRI images are registered to a common coordinate system. Therefore, quality control (QC) of these registrations is necessary to ensure that they are suitable for further processing. Further, in big data studies that involve several thousands of images, manual QC is not feasible and hence there is a need for a fully automated QC mechanism at the pre-processing stage. Checking the quality of rigid and affine registrations is one such task.
The research group implemented a fully automated QC mechanism based on computing several quality metrics local to the image and trained several machine learning classifiers based on these locally computed quality measures. The trained classifiers include linear discriminant analysis, support vector machine, Gaussian naïve Bayes, random forest and adaptive boosting. The developed ML models generalize well to detect misaligned registrations across different MRI scanner vendors and field strengths and even under noisy image situations. Therefore, the classifiers could be employed in big data studies for fully automated QC of registrations, especially T1w and T2w MRI.
Big data MRI studies are generally conducted using a large number of subjects. The conclusions drawn based on big data analysis are more reliable and help to understand the disease mechanics better. The developed method can help to reduce the manual labour for various QC mechanisms required during the pre-processing stage.
This work is done in collaboration with
a. Prof Erik B Dam, Machine Learning Group, University of Copenhagen, Copenhagen, Denmark.
b. Prof Niels K Focke, Clinic for neurology, University Medical Centre, Göttingen, Germany.
In future, the idea is to implement the QC framework using sparse autoencoders in an unsupervised manner and also using Siamese neural networks via deep representation learning.

