Dr Abhijit Dasgupta, Assistant Professor from the Department of Computer Science and Engineering, has published his breakthrough research article titled “Turnover Atlas of Proteome and Phosphoproteome Across Mouse Tissues and Brain Regions” in the nature index journal Cell having an impact factor of 45.5.
Abstract
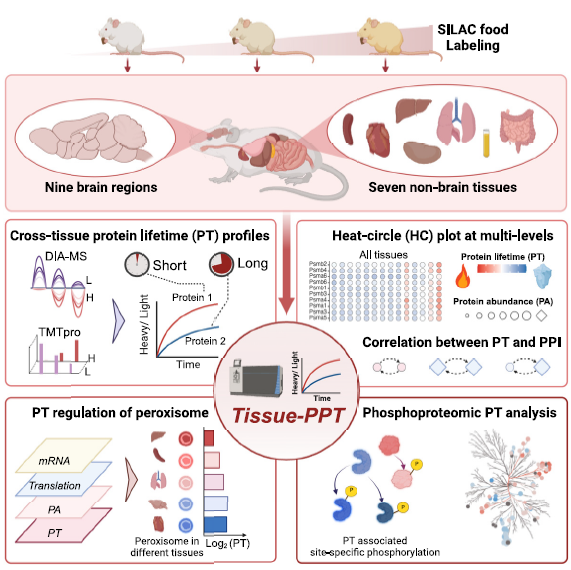
This study presents a comprehensive in vivo atlas of protein and phosphoprotein turnover across 16 mouse tissues and brain regions, integrating advanced mass spectrometry with stable isotope labeling. By mapping over 11,000 proteins and 40,000 phosphosites, the study reveals tissue-specific patterns of protein lifetimes, correlating them with abundance, function, and protein-protein interactions (PPI). It highlights how phosphorylation regulates protein stability and how turnover is linked to peroxisome function, ubiquitination, and neurodegeneration-associated proteins such as Tau and α-synuclein. This high-resolution resource enhances our understanding of proteostasis and dynamic protein regulation, providing new insights into tissue-specific physiology and disease mechanisms.
Explanation of the Research in Layperson’s Terms
All cells in the body continuously make and break down proteins. The balance of these processes—called protein turnover—is vital for keeping tissues healthy. But until now, scientists didn’t have a clear, detailed map of how protein turnover works across different tissues and brain regions.
In this study, researchers used advanced techniques to measure how long thousands of proteins and their phosphorylated (chemically modified) versions last in 16 parts of the mouse body. They discovered that some proteins, especially in the brain and heart, live much longer than others. They also found that proteins interacting with each other often have similar lifespans, and that specific chemical modifications like phosphorylation can either stabilize or destabilize key proteins—such as those involved in Alzheimer’s and Parkinson’s diseases.
The team created an online tool that lets other scientists explore this rich dataset. The findings can help understand tissue function better and may lead to new ways to treat diseases by targeting protein stability.
Practical Implementation/ Social Implications of the Research
Practical Implementation:
This turnover atlas provides a foundational resource for drug development and tissue-specific disease research. It supports AI-driven approaches to predict protein dynamics, aids in identifying long-lived disease-related proteins, like Tau and α-synuclein, and enhances biomarker discovery for neurodegenerative and metabolic diseases. The integrated tool Tissue-PPT allows researchers to explore and analyze protein lifespan and phosphorylation patterns across tissues.
Social Implications:
Understanding how proteins behave differently across tissues could help create more precise therapies for complex diseases such as Alzheimer’s, Parkinson’s, and cardiomyopathies. The dataset empowers researchers globally to explore protein turnover without relying heavily on animal experiments, advancing ethical and efficient biomedical research.
Collaborations
- Yale University School of Medicine, CT, USA
- St. Jude Children’s Research Hospital, TN, USA
- University Medical Center Göttingen, Germany
- University of Trieste, Italy
- West China Hospital, Sichuan University, China
Future Research Plans
The next phase will focus on AI-driven modeling of site-specific phosphorylation turnover in relation to disease phenotypes, using the Tissue-PPT dataset as a foundation. This includes integrating proteomics, phosphoproteomics, and transcriptomic data to refine our understanding of proteome regulation. Special attention will be given to how phosphorylation modulates the stability of neurodegenerative disease proteins and the development of targeted dephosphorylation therapeutics (e.g., PhosTACs).
This research aims to inform personalised interventions and identify novel therapeutic targets by understanding how tissue-specific protein lifespans are regulated under physiological and pathological conditions.